Adjoint gradients for the SPE1 model
Blackoil HistoryMatching Differentiability InputFileThis is a brief example of how to set up a case and compute adjoint gradients for the SPE1 model in a few different ways. These techniques are in general applicable to DATA-type models.
Load the model
using Jutul, JutulDarcy, GeoEnergyIO, GLMakie
data_pth = joinpath(GeoEnergyIO.test_input_file_path("SPE1"), "SPE1.DATA")
data = parse_data_file(data_pth);
case = setup_case_from_data_file(data);Set up a function to set up the case with custom porosity
We create a setup function that takes in a parameter dictionary prm and returns a case with the porosity set to the value in prm["poro"]. This is a convenient way to customize a case prior to setup, allowing direct interaction with keywords like PERMX, PORO, etc.
function F(prm, step_info = missing)
data_c = deepcopy(data)
data_c["GRID"]["PORO"] = fill(prm["poro"], size(data_c["GRID"]["PORO"]))
case = setup_case_from_data_file(data_c)
return case
endF (generic function with 2 methods)Define and simulate truth case (poro from input)
x_truth = only(unique(data["GRID"]["PORO"]))
prm_truth = Dict("poro" => x_truth)
case_truth = F(prm_truth)
ws, states = simulate_reservoir(case_truth)ReservoirSimResult with 120 entries:
wells (2 present):
:INJ
:PROD
Results per well:
:wrat => Vector{Float64} of size (120,)
:Aqueous_mass_rate => Vector{Float64} of size (120,)
:orat => Vector{Float64} of size (120,)
:bhp => Vector{Float64} of size (120,)
:mrat => Vector{Float64} of size (120,)
:gor => Vector{Float64} of size (120,)
:lrat => Vector{Float64} of size (120,)
:mass_rate => Vector{Float64} of size (120,)
:rate => Vector{Float64} of size (120,)
:Vapor_mass_rate => Vector{Float64} of size (120,)
:control => Vector{Symbol} of size (120,)
:Liquid_mass_rate => Vector{Float64} of size (120,)
:wcut => Vector{Float64} of size (120,)
:grat => Vector{Float64} of size (120,)
states (Vector with 120 entries, reservoir variables for each state)
:Pressure => Vector{Float64} of size (300,)
:ImmiscibleSaturation => Vector{Float64} of size (300,)
:BlackOilUnknown => Vector{BlackOilX{Float64}} of size (300,)
:TotalMasses => Matrix{Float64} of size (3, 300)
:Rs => Vector{Float64} of size (300,)
:Saturations => Matrix{Float64} of size (3, 300)
time (report time for each state)
Vector{Float64} of length 120
result (extended states, reports)
SimResult with 120 entries
extra
Dict{Any, Any} with keys :simulator, :config
Completed at Feb. 04 2026 09:17 after 6 seconds, 603 milliseconds, 432.7 microseconds.Define pressure difference as objective function
function pdiff(p, p0)
v = 0.0
for i in eachindex(p)
v += (p[i] - p0[i])^2
end
return v
end
step_times = cumsum(case.dt)
total_time = step_times[end]
function mismatch_objective_p(m, s, dt, step_info, forces)
t = step_info[:time] + dt
step = findmin(x -> abs(x - t), step_times)[2]
p = s[:Reservoir][:Pressure]
v = pdiff(p, states[step][:Pressure])
return (dt/total_time)*(v/(si_unit(:bar)*100)^2)
endmismatch_objective_p (generic function with 1 method)Create a perturbed initial guess and optimize
We create a perturbed initial guess for the porosity and optimize the case using the mismatch objective function defined above. The optimization will adjust the porosity to minimize the mismatch between the simulated pressure and the truth case, and recover the original porosity value of 0.3.
prm = Dict("poro" => x_truth .+ 0.25)
dprm = setup_reservoir_dict_optimization(prm, F)
free_optimization_parameter!(dprm, "poro", abs_max = 1.0, abs_min = 0.1)
prm_opt = optimize_reservoir(dprm, mismatch_objective_p);
dprmDictParameters with 1 parameters (1 active), and 0 multipliers:
Active optimization parameters
┌──────┬───────────────┬───────┬─────┬─────┬─────────────────┬────────┐
│ Name │ Initial value │ Count │ Min │ Max │ Optimized value │ Change │
├──────┼───────────────┼───────┼─────┼─────┼─────────────────┼────────┤
│ poro │ 0.55 │ 1 │ 0.1 │ 1.0 │ 0.295 │ -46.0% │
└──────┴───────────────┴───────┴─────┴─────┴─────────────────┴────────┘
No inactive optimization parameters.
No multipliers set.Plot the optimization history
using GLMakie
fig = Figure()
ax = Axis(fig[1, 1], xlabel = "LBFGS iteration", ylabel = "Objective function", yscale = log10)
scatter!(ax, dprm.history.objectives)
fig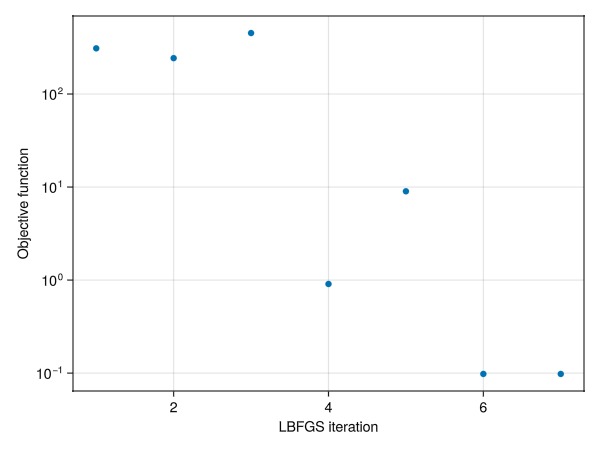
Use lumping to match permeability
The model contains three layers with different permeability. We can perturb the permeability in each layer a bit and use the lumping and scaling functionality in the optimizer to recover the original values.
We first define the setup function which expands a permeability value in each cell to PERMX, PERMY and PERMZ, as these are equal for the base model.
rmesh = physical_representation(reservoir_domain(case.model))
layerno = map(i -> cell_ijk(rmesh, i)[3], 1:number_of_cells(rmesh))
darcy = si_unit(:darcy)
function F_perm(prm, step_info = missing)
data_c = deepcopy(data)
sz = size(data_c["GRID"]["PERMX"])
permxyz = reshape(prm["perm"], sz)
data_c["GRID"]["PERMX"] = permxyz
data_c["GRID"]["PERMY"] = permxyz
data_c["GRID"]["PERMZ"] = permxyz
case = setup_case_from_data_file(data_c)
return case
endF_perm (generic function with 2 methods)Define the starting point
We perturb each layer a bit by multiplying with a constant factor to create a case where the pressure matches.
perm_truth = vec(data["GRID"]["PERMX"])
factors = [1.5, 2.0, 5.0]
prm = Dict(
"perm" => perm_truth.*factors[layerno],
)Dict{String, Vector{Float64}} with 1 entry:
"perm" => [7.40192e-13, 7.40192e-13, 7.40192e-13, 7.40192e-13, 7.40192e-13, 7…Optimize with lumping
We can lump parameters of the same name together. In this case, "perm" has one value per cell. We would like to exploit the fact that we know that there should be one unique value per layer as a prior assumption.
We already have a vector with one entry per cell that defines the layers, so we can use this as the lumping parameter directly.
perm_opt = setup_reservoir_dict_optimization(prm, F_perm)
free_optimization_parameter!(perm_opt, "perm", rel_min = 0.1, rel_max = 10.0, lumping = layerno)
perm_tuned = optimize_reservoir(perm_opt, mismatch_objective_p);
perm_optDictParameters with 1 parameters (1 active), and 0 multipliers:
Active optimization parameters
┌──────┬─────────────────────────────┬───────┬─────────────────────────────┬────
│ Name │ Initial value │ Count │ Min │ ⋯
├──────┼─────────────────────────────┼───────┼─────────────────────────────┼────
│ perm │ 7.4e-13, 9.87e-14, 9.87e-13 │ 3 │ 7.4e-14, 9.87e-15, 9.87e-14 │ 7 ⋯
└──────┴─────────────────────────────┴───────┴─────────────────────────────┴────
3 columns omitted
No inactive optimization parameters.
No multipliers set.Plot the recovered permeability
first_entry = map(i -> findfirst(isequal(i), layerno), 1:3)
kval = [perm_truth[first_entry]..., prm["perm"][first_entry]..., perm_tuned["perm"][first_entry]...]
kval = 1000.0.*kval./si_unit(:darcy)
catval = [1, 2, 3, 1, 2, 3, 1, 2, 3]
group = [1, 1, 1, 2, 2, 2, 3, 3, 3]
colors = Makie.wong_colors()
fig, ax, plt = barplot(catval, kval,
dodge = group,
color = colors[group],
axis = (
xticks = (1:3, ["Layer 1", "Layer 2", "Layer 3"]),
ylabel = "Permeability / md",
title = "Tuned permeability layers"
),
)
labels = ["Truth", "Initial guess", "Optimized"]
elements = [PolyElement(polycolor = colors[i]) for i in 1:length(labels)]
title = "Categories"
Legend(fig[1,2], elements, labels, title)
fig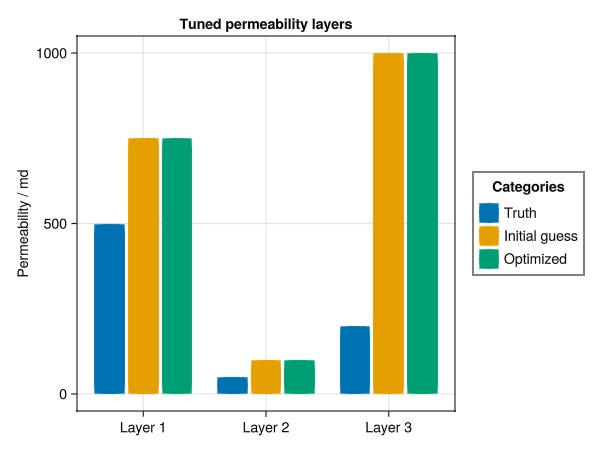
Compute sensitivities outside the optimization
We can also compute the sensitivities outside the optimization process. As our previous setup funciton only has a single parameter (the porosity), we instead switch to the setup_reservoir_dict_optimization function that can set up a set of "typical" tunable parameters for any reservoir model. This saves us the hassle of writing this function ourselves when we want to optimize e.g. permeability, porosity and well indices.
dprm_case = setup_reservoir_dict_optimization(case)
free_optimization_parameters!(dprm_case)
dprm_grad = parameters_gradient_reservoir(dprm_case, mismatch_objective_p);setup_reservoir_state: Received primary variable LiquidSaturation, but this is not known to reservoir model.
setup_reservoir_state: Received primary variable VaporSaturation, but this is not known to reservoir model.
Optimization: Setting up adjoint storage.
Optimization: Finished setup in 3.996821751 seconds.
Optimization: Adjoint solve took 13.96825819 seconds.
Optimization: Objective #1: 4.15354e-01, gradient 2-norm: 3.79058e+14Plot the gradient of the mismatch objective with respect to the porosity
We see, as expected, that the gradient is largest in magnitude around the wells and near the front of the displacement.
m = physical_representation(reservoir_domain(case.model))
fig, ax, plt = plot_cell_data(m, dprm_grad[:model][:porosity])
ax.title[] = "Gradient of mismatch objective with respect to porosity"
fig
Plot the sensitivities in the interactive viewer
If you are running the example yourself, you can now explore the sensitivities in the interactive viewer. This is useful for understanding how the model responds to changes in the parameters.
plot_reservoir(case.model, dprm_grad[:model])
Example on GitHub
If you would like to run this example yourself, it can be downloaded from the JutulDarcy.jl GitHub repository as a script, or as a Jupyter Notebook
This example took 149.341488209 seconds to complete.This page was generated using Literate.jl.